Bacterial translation differs from eukaryotic translation in several ways. Notably, bacterial ribosomes are smaller (70S vs. 80S), use a distinct initiator tRNA and eIF complex, and exhibit polysomes for rapid protein synthesis. Additionally, bacterial mRNA has a Shine-Dalgarno sequence for ribosome binding and exhibits specific codon usage patterns. Moreover, bacteria and eukaryotes employ different sets of translation factors, rendering bacteria susceptible to antibiotics that target translation. These distinctions reflect the unique adaptation of bacterial translation to specific microbial needs and physiological contexts.
Initiation: Divergent Mechanisms in Bacterial and Eukaryotic Translation
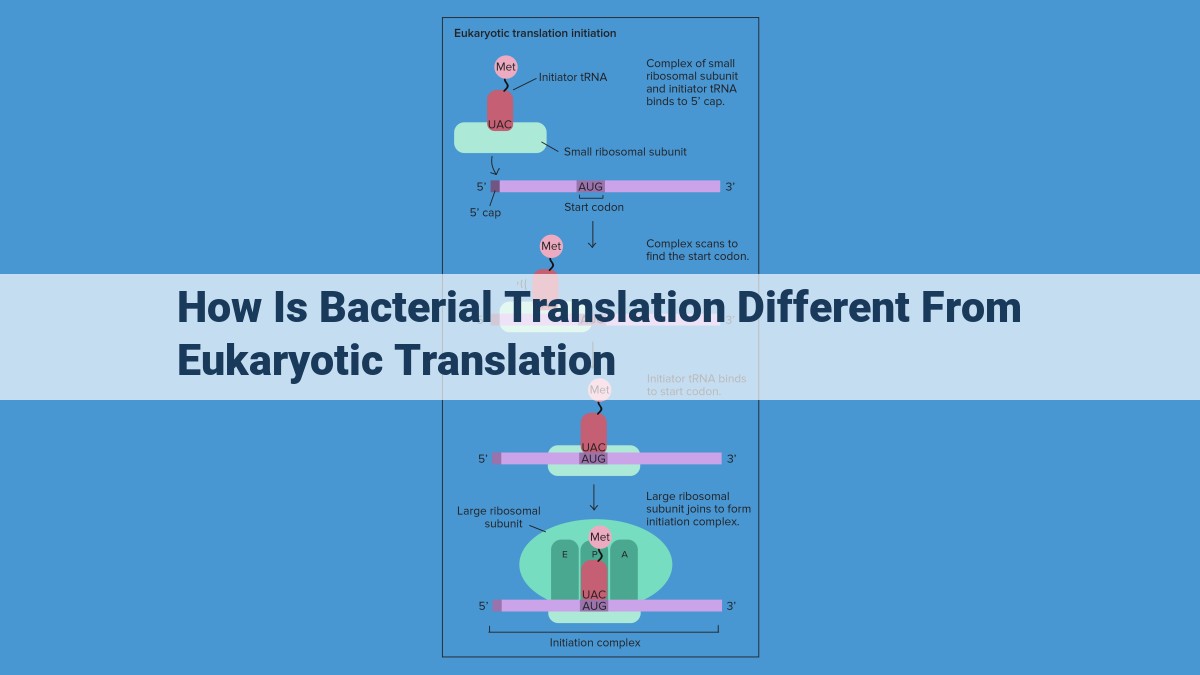
Step into the intricate dance of translation, where genetic code transforms into the proteins that drive cellular life. It’s a journey that begins with initiation, the crucial first step in protein synthesis. But hold on, because the path taken by bacteria and eukaryotes in this dance diverges significantly.
In bacteria, the initiation waltz starts with a specialized tRNA, tailor-made to recognize the start codon (often AUG). It’s like a perfect key fitting into a specific lock. This tRNA is guided by a unique set of initiation factors (eIF), the conductors of the translation orchestra. These eIFs unveil the mRNA’s hidden message, recognizing the ribosome binding site (RBS) and placing the tRNA in the spotlight.
Eukaryotes, on the other hand, have evolved a more elaborate initiation sequence. Their tRNA is also tailored to the start codon, but the eIF family is more extensive and complex. These eIFs carefully scan the mRNA, searching for the cap (a distinctive molecular flag) and the Kozak sequence (a consensus sequence near the start codon). Once these landmarks are identified, the eIFs orchestrate a precise assembly of the ribosome, tRNA, and mRNA, initiating the translation process.
So, while the goal of initiation remains the same for bacteria and eukaryotes – to set the stage for protein synthesis – the mechanisms they employ are as distinct as two sides of a molecular coin.
Ribosomal Differences: Size and Structure
- Explain the smaller size of bacterial 70S ribosomes compared to eukaryotic 80S ribosomes, highlighting the differences in subunit composition and functions.
Ribosomal Differences: Size and Structure
In the world of protein synthesis, ribosomes play the starring role. These molecular machines, responsible for translating genetic code into the proteins that drive our cells, come in two distinct flavors: bacterial 70S ribosomes and eukaryotic 80S ribosomes.
Size Matters: 70S vs. 80S
As their names suggest, bacterial ribosomes are smaller than their eukaryotic counterparts, with a molecular weight of around 70S (Svedberg units). In comparison, eukaryotic ribosomes weigh in at 80S. This difference in size reflects the varying complexity of the organisms they serve.
Subunit Composition: A Tale of Two RNAs
Ribosomes are composed of two subunits: a large subunit and a small subunit, each containing a blend of ribosomal RNA (rRNA) and proteins. In bacteria, the large subunit contains three rRNA molecules (23S, 5S, and 5.8S), while the small subunit contains one rRNA molecule (16S).
In eukaryotes, the large subunit contains three rRNA molecules (28S, 5.8S, and 5S), and the small subunit contains four rRNA molecules (18S, 5.8S, 28S, and 5S). The additional rRNA and proteins in eukaryotic ribosomes contribute to their increased size and complexity.
Functional Differences: Protein Complexity
The large subunit of the ribosome is responsible for peptide bond formation, linking amino acids together to form proteins. The presence of extra rRNA and proteins in eukaryotic large subunits allows them to handle more complex protein structures, such as those requiring extensive disulfide bond formation.
The small subunit of the ribosome scans mRNA, the genetic template for protein synthesis, and binds to the start codon to initiate translation. The larger size of eukaryotic small subunits and the presence of additional proteins provide greater accuracy and versatility in this critical step.
In summary, the smaller size and simpler composition of bacterial 70S ribosomes make them better suited for the rapid protein synthesis required by bacteria. Conversely, the larger size and increased complexity of eukaryotic 80S ribosomes allow them to handle the more demanding protein synthesis processes found in eukaryotic organisms.
Polysomes: Powerhouses of Protein Production
In the bacterial kingdom, where time is of the essence and survival often hinges on the production of essential proteins, a remarkable adaptation has evolved: polysomes. These intricate structures are composed of multiple ribosomes tethered together like a string of pearls, each ribosome diligently translating the same messenger RNA (mRNA) molecule. This ingenious arrangement allows bacteria to massively amplify their protein production capacity.
Unlike their eukaryotic counterparts, which typically have only one ribosome bound to each mRNA molecule, bacteria can form polysomes, which can consist of up to 100 ribosomes simultaneously translating the same genetic blueprint. This extraordinary ribosome density enables bacteria to produce proteins at an astonishing rate, speeding up their response to environmental cues or threats.
The formation of polysomes is meticulously orchestrated by specific proteins that bind to the mRNA and guide multiple ribosomes to the starting point of translation. Once assembled, the polysome becomes a protein synthesis powerhouse, with each ribosome churning out its own copy of the protein encoded by the mRNA.
This efficient protein production system is particularly advantageous for bacteria that need to respond quickly to changing conditions, such as nutrient availability or the presence of predators. It allows them to rapidly synthesize specific proteins that aid in their survival and adaptability.
In essence, polysomes are a testament to the prodigious ingenuity of the bacterial world, providing a powerful mechanism for protein production that is essential for their survival and success in diverse environments.
Codon Usage: The Language of Protein Synthesis
Every cell in our body reads a genetic code, a set of molecular instructions that determine the proteins it will produce. This code is written in a language of codons, three-nucleotide sequences that specify which amino acid will be added to a growing protein chain. While the genetic code is universal across all living organisms, different organisms exhibit distinct preferences for certain codons. This phenomenon is known as codon usage bias.
Bacterial vs. Eukaryotic Codon Usage:
Bacteria and eukaryotes, the two main groups of cells, differ markedly in their codon usage patterns. Bacteria favor codons with a high GC content, while eukaryotes show a preference for codons rich in A and U. These discrepancies reflect the different environments in which these organisms live and the specific proteins they require.
GC-rich Codons in Bacteria:
Bacteria thrive in diverse environments, including extreme conditions such as high temperatures and acidity. GC-rich codons are more stable under these conditions, helping to protect the genetic code from environmental stress. Moreover, GC-rich codons tend to be recognized by specific transfer RNAs (tRNAs), molecules that carry amino acids to the ribosome during protein synthesis. The abundance of these tRNAs in bacteria facilitates rapid protein production.
AU-rich Codons in Eukaryotes:
Eukaryotes, which include plants, animals, and fungi, reside primarily in more stable environments. They have developed a preference for AU-rich codons. These codons are recognized by a wider range of tRNAs, ensuring that the necessary amino acids are readily available for protein synthesis. Additionally, AU-rich codons are more efficiently translated, contributing to the efficient production of complex proteins required for eukaryotic life.
Implications for Disease and Drug Development:
Understanding codon usage bias has significant implications for understanding disease and developing drugs. For example, viruses evolve to use codons that match the host’s preferences, making it easier for them to infect and replicate. Conversely, antibiotics specifically target codons that are highly used by bacteria but not by eukaryotes, reducing the risk of harm to the host.
Codon usage bias is a fascinating aspect of genetics that reflects the diverse evolutionary paths of different organisms. By understanding these preferences, scientists can gain insights into the biology of disease and develop more effective therapies.
**Shine-Dalgarno Sequence: Guiding the Ribosome to the Start Codon in Bacteria**
In the realm of molecular biology, the Shine-Dalgarno sequence plays a pivotal role in the translation process in bacteria. Unlike its eukaryotic counterparts, bacterial mRNA lacks a distinct start codon. Instead, the Shine-Dalgarno sequence, a ribosome-binding site, guides the ribosome to the start codon.
This sequence is typically located 5-10 nucleotides upstream of the start codon, and its complementarity to the anti-Shine-Dalgarno sequence on the 3′ end of the 16S rRNA of the small ribosomal subunit facilitates the ribosome’s precise binding. The strength of the binding between the two sequences influences the efficiency of translation initiation.
The Shine-Dalgarno sequence varies in length, with the most common being AGGAGG (known as the SD sequence). However, different bacterial species may have unique Shine-Dalgarno sequences that are optimized for their specific genetic context.
In eukaryotes, the start codon (AUG) is recognized by the cap structure at the 5′ end of the mRNA and the Kozak sequence (GCCRCCAUGG), which is located immediately upstream of the start codon. This more complex start codon recognition mechanism is not required in bacteria due to the presence of the Shine-Dalgarno sequence.
Conclusion: The Shine-Dalgarno sequence is an essential feature of bacterial mRNA that facilitates ribosome binding and start codon recognition. Its absence in eukaryotes underscores the distinct mechanisms of translation initiation in these two domains of life.
Translation Factors: Molecular Facilitators of Protein Synthesis
Like skilled choreographers, translation factors play a vital role in the complex dance of protein synthesis, guiding the ribosome through the intricate steps of translation. These molecular maestros possess distinct roles in both bacteria and eukaryotes, each orchestrating the assembly and disassembly of the ribosomal machinery with remarkable precision.
Bacterial Translation Factors: A Streamlined Symphony
Bacteria employ a leaner team of translation factors than their eukaryotic counterparts. Initiation factor 3 (IF3) orchestrates the assembly of the initiation complex, pairing the small ribosomal subunit with the messenger RNA (mRNA) and the initiator tRNA. Once this partnership is forged, IF2 chaperones the large ribosomal subunit into place, completing the 70S initiation complex.
Elongation factor Tu (EF-Tu) is the tireless workhorse of bacterial translation, shepherding aminoacyl-tRNAs to the growing polypeptide chain. Each new amino acid is meticulously added by elongation factor G (EF-G), which drives the ribosome along the mRNA with unwavering efficiency.
Finally, release factors RF1 and RF2 gracefully end the translation process, signaling the termination of protein synthesis and the release of the newly synthesized polypeptide into the cellular landscape.
Eukaryotic Translation Factors: A Grand Orchestra
Eukaryotic translation factors form a far more elaborate ensemble. The initiation process is spearheaded by eukaryotic initiation factor 4E (eIF4E), which binds the mRNA cap and recruits other initiation factors to the fray. eIF3 assembles the small ribosomal subunit, while eIF2 recruits the initiator tRNA and pairs it with the start codon. After the large ribosomal subunit joins the complex, eIF5 orchestrates the final assembly steps.
eEF1A and eEF2 collaborate seamlessly in the elongation phase, delivering aminoacyl-tRNAs and catalyzing the formation of peptide bonds. Once the ribosome reaches the stop codon, release factors eRF1 and eRF3 dissolve the translation complex and liberate the newly synthesized protein.
Antibiotic Susceptibility: Targeting Bacterial Translation
The intricate machinery of bacterial translation has become a prime target for antibiotic therapy. Drugs like tetracycline and erythromycin cripple the function of EF-Tu, disrupting the delivery of amino acids and stunting protein synthesis. Chloramphenicol also targets EF-Tu, blocking the transfer of aminoacyl-tRNAs to the ribosome. By interfering with these essential translation factors, antibiotics effectively halt bacterial growth without affecting eukaryotic protein synthesis, making them invaluable weapons in the fight against bacterial infections.
Antibiotic Susceptibility: Targeting the Achilles’ Heel of Bacterial Translation
Bacteria, the formidable microorganisms that can cause a myriad of infections, rely heavily on their ability to translate genetic information into essential proteins. However, this crucial process harbors a critical vulnerability that humans have ingeniously exploited to develop antibiotics. These wonder drugs selectively target bacterial translation, crippling the bacteria’s ability to produce life-sustaining proteins without harming human cells.
Antibiotics achieve this feat by disrupting specific steps in the bacterial translation process. For instance, tetracycline inhibits the binding of tRNA to the ribosome, the cellular machinery responsible for protein synthesis. Another antibiotic, chloramphenicol, blocks the elongation of the growing polypeptide chain. By effectively halting protein production, these antibiotics effectively cripple bacterial growth and multiplication.
The remarkable specificity of antibiotics towards bacterial translation stems from the fundamental differences between bacterial and eukaryotic ribosomes. Bacterial ribosomes, smaller and simpler than their eukaryotic counterparts, exhibit unique structural features that create ideal binding sites for antibiotics. Conversely, eukaryotic ribosomes possess a more complex structure and lack these vulnerable sites, rendering them immune to these antimicrobials.
Antibiotic susceptibility has played a pivotal role in the fight against bacterial infections. Antibiotics have revolutionized medicine, allowing us to effectively treat once-fatal diseases such as pneumonia, tuberculosis, and meningitis. However, the widespread use of antibiotics has also fueled the emergence of antibiotic resistance, a formidable threat to global health.
To overcome this challenge, researchers are actively pursuing novel antibiotic targets that exploit other vulnerabilities in bacterial translation. By understanding the intricacies of this fundamental process, scientists hope to develop new and effective antibiotics to combat the growing menace of antibiotic resistance.